Author: PuTI
Mass importing .txt files with sequential names to plot multiple graphs on same figure for easy comparison
I have 30 .txt files numbered like so:
FreeField1.txt
FreeField2.txt
…
FreeField30.txt
I need to find a way to import the data from all the files at once so I can then plot a graph with all of them overlaid to allow for quick comparison.
The current method I have found which inputs all the data is like so
numfiles = 30;
S = dir(‘*.txt’)
N = length(S)
for i = 1:numel(S)
S(i).data = readtable(S(i).name)
end
But this gives a structure which I do not how to progress from. I am looking for any possible methods which could be used for this mass importation to then build off for the plotting of graphs.
Any help would be appreciated as I am very new to MATLab and am wishing to understand it more. I have attached a sample text file for convenience.I have 30 .txt files numbered like so:
FreeField1.txt
FreeField2.txt
…
FreeField30.txt
I need to find a way to import the data from all the files at once so I can then plot a graph with all of them overlaid to allow for quick comparison.
The current method I have found which inputs all the data is like so
numfiles = 30;
S = dir(‘*.txt’)
N = length(S)
for i = 1:numel(S)
S(i).data = readtable(S(i).name)
end
But this gives a structure which I do not how to progress from. I am looking for any possible methods which could be used for this mass importation to then build off for the plotting of graphs.
Any help would be appreciated as I am very new to MATLab and am wishing to understand it more. I have attached a sample text file for convenience. I have 30 .txt files numbered like so:
FreeField1.txt
FreeField2.txt
…
FreeField30.txt
I need to find a way to import the data from all the files at once so I can then plot a graph with all of them overlaid to allow for quick comparison.
The current method I have found which inputs all the data is like so
numfiles = 30;
S = dir(‘*.txt’)
N = length(S)
for i = 1:numel(S)
S(i).data = readtable(S(i).name)
end
But this gives a structure which I do not how to progress from. I am looking for any possible methods which could be used for this mass importation to then build off for the plotting of graphs.
Any help would be appreciated as I am very new to MATLab and am wishing to understand it more. I have attached a sample text file for convenience. data import, graph, text file MATLAB Answers — New Questions
3D surf plot for more than two quantities
I want to plot the value in the same 3D graph .
syms x t r b %alpha
% Parameter values
a=(pi)/3;
g=9.8;
U=2.5;
O=7.29*10^(-5);
f=2*O*sin(a);
H=-(f/g)*U;
alpha=0.75; % fractional order
%%%%%%%%%initalization of variable
u_l=sym(zeros(1));
v_l=zeros(1,’sym’);
h_l=zeros(1,’sym’);
A_l=zeros(1,2,’sym’);
B_l=zeros(1,2,’sym’);
C_l=zeros(1,2,’sym’);
D_l=zeros(1,2,’sym’);
series1_l(x,t)=sym(zeros(1,1));
series2_l(x,t)=sym(zeros(1,1));
series3_l(x,t)=sym(zeros(1,1));
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
u_up=sym(zeros(1));
v_up=zeros(1,’sym’);
h_up=zeros(1,’sym’);
A_up=zeros(1,2,’sym’);
B_up=zeros(1,2,’sym’);
C_up=zeros(1,2,’sym’);
D_up=zeros(1,2,’sym’);
series1_up(x,t)=sym(zeros(1,1));
series2_up(x,t)=sym(zeros(1,1));
series3_up(x,t)=sym(zeros(1,1));
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
u_cr=sym(zeros(1));
v_cr=zeros(1,’sym’);
h_cr=zeros(1,’sym’);
A_cr=zeros(1,2,’sym’);
B_cr=zeros(1,2,’sym’);
C_cr=zeros(1,2,’sym’);
D_cr=zeros(1,2,’sym’);
series1_cr(x,t)=sym(zeros(1,1));
series2_cr(x,t)=sym(zeros(1,1));
series3_cr(x,t)=sym(zeros(1,1));
%%%%%%%% Initial condition fuzzy condition
R=0.5;
b_l=0 % lower bound
b_cr=0.5 % middle value
b_u=1 % upper value
%%%%%%%%%%%%%%%%%% lOWER VALUE%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
u_l(1)=(2*b_l*(1-R)+R)*exp(x)*(sech(x))^2;
v_l(1)=(2*b_l*(1-R)+R+1)*2*x*(sech(2*x))^2;
h_l(1)=(2*b_l*(1-R)+R)*x^2*(sech(2*x))^2;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%UPPER VALUE
u_up(1)=(2*b_u*(1-R)+R)*exp(x)*(sech(x))^2;
v_up(1)=(2*b_u*(1-R)+R+1)*2*x*(sech(2*x))^2;
h_up(1)=(2*b_u*(1-R)+R)*x^2*(sech(2*x))^2;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%Middle valur %%%%%%%%%%%%%%%%%%%%
u_cr(1)=(2*b_cr*(1-R)+R)*exp(x)*(sech(x))^2;
v_cr(1)=(2*b_cr*(1-R)+R+1)*2*x*(sech(2*x))^2;
h_cr(1)=(2*b_cr*(1-R)+R)*x^2*(sech(2*x))^2;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%lOWER VALUR %%%%%%%%%%%%%%%%%%%%%%%%%
for k=1:2
A_l=0;
B_l=0;
C_l=0;
D_l=0;
if k==1
E=1;
else
E=0;
end
for r=1:k
A_l=A_l+u_l(r)*diff(u_l(k-r+1),x,1);
B_l=B_l+u_l(r)*diff(v_l(k-r+1),x,1);
C_l=C_l+u_l(r)*diff(h_l(k-r+1),x,1);
D_l=D_l+h_l(r)*diff(u_l(k-r+1),x,1);
end
u_l(k+1)=simplify(gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-A_l+f*v_l(k)-g*diff(h_l(k),x,1)));
v_l(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-B_l-f*u_l(k)-g*H*E);
h_l(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-C_l-H*v_l(k)-D_l);
end
% t1=simplify(u(2))
% var2 = vpa(t1)
% t2=simplify(v(2))
% var = vpa(t2)
% t3=simplify(h(2))
for k=1:3
series1_l(x,t)=simplify(series1_l(x,t)+u_l(k)*(power(t,k-1)));
series2_l(x,t)=simplify(series2_l(x,t)+v_l(k)*(power(t,k-1)));
series3_l(x,t)=simplify(series3_l(x,t)+h_l(k)*(power(t,k-1)));
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% UPPER VALUE %%%%%%%%%%%%%%%%
for k=1:2
A_up=0;
B_up=0;
C_up=0;
D_up=0;
if k==1
E=1;
else
E=0;
end
for r=1:k
A_up=A_up+u_up(r)*diff(u_up(k-r+1),x,1);
B_up=B_up+u_up(r)*diff(v_up(k-r+1),x,1);
C_up=C_up+u_up(r)*diff(h_up(k-r+1),x,1);
D_up=D_up+h_up(r)*diff(u_up(k-r+1),x,1);
end
u_up(k+1)=simplify(gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-A_up+f*v_up(k)-g*diff(h_up(k),x,1)));
v_up(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-B_up-f*u_up(k)-g*H*E);
h_up(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-C_up-H*v_up(k)-D_up);
end
% t1=simplify(u(2))
% var2 = vpa(t1)_u
% t2=simplify(v(2))
% var = vpa(t2)
% t3=simplify(h(2))
for k=1:3
series1_up(x,t)=simplify(series1_up(x,t)+u_up(k)*(power(t,k-1)));
series2_up(x,t)=simplify(series2_up(x,t)+v_up(k)*(power(t,k-1)));
series3_up(x,t)=simplify(series3_up(x,t)+h_up(k)*(power(t,k-1)));
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%middle value %%%%%%%%%%%%%%
for k=1:2
A_cr=0;
B_cr=0;
C_cr=0;
D_cr=0;
if k==1
E=1;
else
E=0;
end
for r=1:k
A_cr=A_cr+u_cr(r)*diff(u_up(k-r+1),x,1);
B_cr=B_cr+u_cr(r)*diff(v_up(k-r+1),x,1);
C_cr=C_cr+u_cr(r)*diff(h_up(k-r+1),x,1);
D_cr=D_up+h_cr(r)*diff(u_up(k-r+1),x,1);
end
u_cr(k+1)=simplify(gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-A_cr+f*v_up(k)-g*diff(h_cr(k),x,1)));
v_cr(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-B_cr-f*u_cr(k)-g*H*E);
h_cr(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-C_cr-H*v_cr(k)-D_cr);
end
% t1=simplify(u(2))
% var2 = vpa(t1)_u
% t2=simplify(v(2))
% var = vpa(t2)
% t3=simplify(h(2))
for k=1:3
series1_cr(x,t)=simplify(series1_cr(x,t)+u_cr(k)*(power(t,k-1)));
series2_cr(x,t)=simplify(series2_cr(x,t)+v_cr(k)*(power(t,k-1)));
series3_cr(x,t)=simplify(series3_cr(x,t)+h_cr(k)*(power(t,k-1)));
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
C1_l=zeros(1)
C2_l=zeros(1)
C3_l=zeros(1)
C1_up=zeros(1)
C2_up=zeros(1)
C3_up=zeros(1)
C1_cr=zeros(1)
C2_cr=zeros(1)
C3_cr=zeros(1)
row=0;
x=0:0.04:2
t=0:0.002:0.1
for i=1:length(x)
row=row+1;
col=0;
for j=1:length(t)
col=col+1;
C1_l(row,col)=series1_l(x(i),t(j));
C2_l(row,col)=series2_l(x(i),t(j));
C3_l(row,col)=series3_l(x(i),t(j));
%———————————-
C1_up(row,col)=series1_up(x(i),t(j));
C2_up(row,col)=series2_up(x(i),t(j));
C3_up(row,col)=series3_up(x(i),t(j));
%————————————
C1_cr(row,col)=series1_cr(x(i),t(j));
C2_cr(row,col)=series2_cr(x(i),t(j));
C3_cr(row,col)=series3_cr(x(i),t(j));
end
end
%——————————————————–
surf(x,t,C1_l,C1_up,C1_cr)
surf(x,t,C2_l,C2_up,C2_cr)
surf(x,t,C3_l,C3_up,C3_cr)
I want to plot all the graph in the same 3D plot. I dont want different graph using subplot. and I am using MATLAB version 2023aI want to plot the value in the same 3D graph .
syms x t r b %alpha
% Parameter values
a=(pi)/3;
g=9.8;
U=2.5;
O=7.29*10^(-5);
f=2*O*sin(a);
H=-(f/g)*U;
alpha=0.75; % fractional order
%%%%%%%%%initalization of variable
u_l=sym(zeros(1));
v_l=zeros(1,’sym’);
h_l=zeros(1,’sym’);
A_l=zeros(1,2,’sym’);
B_l=zeros(1,2,’sym’);
C_l=zeros(1,2,’sym’);
D_l=zeros(1,2,’sym’);
series1_l(x,t)=sym(zeros(1,1));
series2_l(x,t)=sym(zeros(1,1));
series3_l(x,t)=sym(zeros(1,1));
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
u_up=sym(zeros(1));
v_up=zeros(1,’sym’);
h_up=zeros(1,’sym’);
A_up=zeros(1,2,’sym’);
B_up=zeros(1,2,’sym’);
C_up=zeros(1,2,’sym’);
D_up=zeros(1,2,’sym’);
series1_up(x,t)=sym(zeros(1,1));
series2_up(x,t)=sym(zeros(1,1));
series3_up(x,t)=sym(zeros(1,1));
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
u_cr=sym(zeros(1));
v_cr=zeros(1,’sym’);
h_cr=zeros(1,’sym’);
A_cr=zeros(1,2,’sym’);
B_cr=zeros(1,2,’sym’);
C_cr=zeros(1,2,’sym’);
D_cr=zeros(1,2,’sym’);
series1_cr(x,t)=sym(zeros(1,1));
series2_cr(x,t)=sym(zeros(1,1));
series3_cr(x,t)=sym(zeros(1,1));
%%%%%%%% Initial condition fuzzy condition
R=0.5;
b_l=0 % lower bound
b_cr=0.5 % middle value
b_u=1 % upper value
%%%%%%%%%%%%%%%%%% lOWER VALUE%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
u_l(1)=(2*b_l*(1-R)+R)*exp(x)*(sech(x))^2;
v_l(1)=(2*b_l*(1-R)+R+1)*2*x*(sech(2*x))^2;
h_l(1)=(2*b_l*(1-R)+R)*x^2*(sech(2*x))^2;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%UPPER VALUE
u_up(1)=(2*b_u*(1-R)+R)*exp(x)*(sech(x))^2;
v_up(1)=(2*b_u*(1-R)+R+1)*2*x*(sech(2*x))^2;
h_up(1)=(2*b_u*(1-R)+R)*x^2*(sech(2*x))^2;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%Middle valur %%%%%%%%%%%%%%%%%%%%
u_cr(1)=(2*b_cr*(1-R)+R)*exp(x)*(sech(x))^2;
v_cr(1)=(2*b_cr*(1-R)+R+1)*2*x*(sech(2*x))^2;
h_cr(1)=(2*b_cr*(1-R)+R)*x^2*(sech(2*x))^2;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%lOWER VALUR %%%%%%%%%%%%%%%%%%%%%%%%%
for k=1:2
A_l=0;
B_l=0;
C_l=0;
D_l=0;
if k==1
E=1;
else
E=0;
end
for r=1:k
A_l=A_l+u_l(r)*diff(u_l(k-r+1),x,1);
B_l=B_l+u_l(r)*diff(v_l(k-r+1),x,1);
C_l=C_l+u_l(r)*diff(h_l(k-r+1),x,1);
D_l=D_l+h_l(r)*diff(u_l(k-r+1),x,1);
end
u_l(k+1)=simplify(gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-A_l+f*v_l(k)-g*diff(h_l(k),x,1)));
v_l(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-B_l-f*u_l(k)-g*H*E);
h_l(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-C_l-H*v_l(k)-D_l);
end
% t1=simplify(u(2))
% var2 = vpa(t1)
% t2=simplify(v(2))
% var = vpa(t2)
% t3=simplify(h(2))
for k=1:3
series1_l(x,t)=simplify(series1_l(x,t)+u_l(k)*(power(t,k-1)));
series2_l(x,t)=simplify(series2_l(x,t)+v_l(k)*(power(t,k-1)));
series3_l(x,t)=simplify(series3_l(x,t)+h_l(k)*(power(t,k-1)));
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% UPPER VALUE %%%%%%%%%%%%%%%%
for k=1:2
A_up=0;
B_up=0;
C_up=0;
D_up=0;
if k==1
E=1;
else
E=0;
end
for r=1:k
A_up=A_up+u_up(r)*diff(u_up(k-r+1),x,1);
B_up=B_up+u_up(r)*diff(v_up(k-r+1),x,1);
C_up=C_up+u_up(r)*diff(h_up(k-r+1),x,1);
D_up=D_up+h_up(r)*diff(u_up(k-r+1),x,1);
end
u_up(k+1)=simplify(gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-A_up+f*v_up(k)-g*diff(h_up(k),x,1)));
v_up(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-B_up-f*u_up(k)-g*H*E);
h_up(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-C_up-H*v_up(k)-D_up);
end
% t1=simplify(u(2))
% var2 = vpa(t1)_u
% t2=simplify(v(2))
% var = vpa(t2)
% t3=simplify(h(2))
for k=1:3
series1_up(x,t)=simplify(series1_up(x,t)+u_up(k)*(power(t,k-1)));
series2_up(x,t)=simplify(series2_up(x,t)+v_up(k)*(power(t,k-1)));
series3_up(x,t)=simplify(series3_up(x,t)+h_up(k)*(power(t,k-1)));
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%middle value %%%%%%%%%%%%%%
for k=1:2
A_cr=0;
B_cr=0;
C_cr=0;
D_cr=0;
if k==1
E=1;
else
E=0;
end
for r=1:k
A_cr=A_cr+u_cr(r)*diff(u_up(k-r+1),x,1);
B_cr=B_cr+u_cr(r)*diff(v_up(k-r+1),x,1);
C_cr=C_cr+u_cr(r)*diff(h_up(k-r+1),x,1);
D_cr=D_up+h_cr(r)*diff(u_up(k-r+1),x,1);
end
u_cr(k+1)=simplify(gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-A_cr+f*v_up(k)-g*diff(h_cr(k),x,1)));
v_cr(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-B_cr-f*u_cr(k)-g*H*E);
h_cr(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-C_cr-H*v_cr(k)-D_cr);
end
% t1=simplify(u(2))
% var2 = vpa(t1)_u
% t2=simplify(v(2))
% var = vpa(t2)
% t3=simplify(h(2))
for k=1:3
series1_cr(x,t)=simplify(series1_cr(x,t)+u_cr(k)*(power(t,k-1)));
series2_cr(x,t)=simplify(series2_cr(x,t)+v_cr(k)*(power(t,k-1)));
series3_cr(x,t)=simplify(series3_cr(x,t)+h_cr(k)*(power(t,k-1)));
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
C1_l=zeros(1)
C2_l=zeros(1)
C3_l=zeros(1)
C1_up=zeros(1)
C2_up=zeros(1)
C3_up=zeros(1)
C1_cr=zeros(1)
C2_cr=zeros(1)
C3_cr=zeros(1)
row=0;
x=0:0.04:2
t=0:0.002:0.1
for i=1:length(x)
row=row+1;
col=0;
for j=1:length(t)
col=col+1;
C1_l(row,col)=series1_l(x(i),t(j));
C2_l(row,col)=series2_l(x(i),t(j));
C3_l(row,col)=series3_l(x(i),t(j));
%———————————-
C1_up(row,col)=series1_up(x(i),t(j));
C2_up(row,col)=series2_up(x(i),t(j));
C3_up(row,col)=series3_up(x(i),t(j));
%————————————
C1_cr(row,col)=series1_cr(x(i),t(j));
C2_cr(row,col)=series2_cr(x(i),t(j));
C3_cr(row,col)=series3_cr(x(i),t(j));
end
end
%——————————————————–
surf(x,t,C1_l,C1_up,C1_cr)
surf(x,t,C2_l,C2_up,C2_cr)
surf(x,t,C3_l,C3_up,C3_cr)
I want to plot all the graph in the same 3D plot. I dont want different graph using subplot. and I am using MATLAB version 2023a I want to plot the value in the same 3D graph .
syms x t r b %alpha
% Parameter values
a=(pi)/3;
g=9.8;
U=2.5;
O=7.29*10^(-5);
f=2*O*sin(a);
H=-(f/g)*U;
alpha=0.75; % fractional order
%%%%%%%%%initalization of variable
u_l=sym(zeros(1));
v_l=zeros(1,’sym’);
h_l=zeros(1,’sym’);
A_l=zeros(1,2,’sym’);
B_l=zeros(1,2,’sym’);
C_l=zeros(1,2,’sym’);
D_l=zeros(1,2,’sym’);
series1_l(x,t)=sym(zeros(1,1));
series2_l(x,t)=sym(zeros(1,1));
series3_l(x,t)=sym(zeros(1,1));
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
u_up=sym(zeros(1));
v_up=zeros(1,’sym’);
h_up=zeros(1,’sym’);
A_up=zeros(1,2,’sym’);
B_up=zeros(1,2,’sym’);
C_up=zeros(1,2,’sym’);
D_up=zeros(1,2,’sym’);
series1_up(x,t)=sym(zeros(1,1));
series2_up(x,t)=sym(zeros(1,1));
series3_up(x,t)=sym(zeros(1,1));
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
u_cr=sym(zeros(1));
v_cr=zeros(1,’sym’);
h_cr=zeros(1,’sym’);
A_cr=zeros(1,2,’sym’);
B_cr=zeros(1,2,’sym’);
C_cr=zeros(1,2,’sym’);
D_cr=zeros(1,2,’sym’);
series1_cr(x,t)=sym(zeros(1,1));
series2_cr(x,t)=sym(zeros(1,1));
series3_cr(x,t)=sym(zeros(1,1));
%%%%%%%% Initial condition fuzzy condition
R=0.5;
b_l=0 % lower bound
b_cr=0.5 % middle value
b_u=1 % upper value
%%%%%%%%%%%%%%%%%% lOWER VALUE%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
u_l(1)=(2*b_l*(1-R)+R)*exp(x)*(sech(x))^2;
v_l(1)=(2*b_l*(1-R)+R+1)*2*x*(sech(2*x))^2;
h_l(1)=(2*b_l*(1-R)+R)*x^2*(sech(2*x))^2;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%UPPER VALUE
u_up(1)=(2*b_u*(1-R)+R)*exp(x)*(sech(x))^2;
v_up(1)=(2*b_u*(1-R)+R+1)*2*x*(sech(2*x))^2;
h_up(1)=(2*b_u*(1-R)+R)*x^2*(sech(2*x))^2;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%Middle valur %%%%%%%%%%%%%%%%%%%%
u_cr(1)=(2*b_cr*(1-R)+R)*exp(x)*(sech(x))^2;
v_cr(1)=(2*b_cr*(1-R)+R+1)*2*x*(sech(2*x))^2;
h_cr(1)=(2*b_cr*(1-R)+R)*x^2*(sech(2*x))^2;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%lOWER VALUR %%%%%%%%%%%%%%%%%%%%%%%%%
for k=1:2
A_l=0;
B_l=0;
C_l=0;
D_l=0;
if k==1
E=1;
else
E=0;
end
for r=1:k
A_l=A_l+u_l(r)*diff(u_l(k-r+1),x,1);
B_l=B_l+u_l(r)*diff(v_l(k-r+1),x,1);
C_l=C_l+u_l(r)*diff(h_l(k-r+1),x,1);
D_l=D_l+h_l(r)*diff(u_l(k-r+1),x,1);
end
u_l(k+1)=simplify(gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-A_l+f*v_l(k)-g*diff(h_l(k),x,1)));
v_l(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-B_l-f*u_l(k)-g*H*E);
h_l(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-C_l-H*v_l(k)-D_l);
end
% t1=simplify(u(2))
% var2 = vpa(t1)
% t2=simplify(v(2))
% var = vpa(t2)
% t3=simplify(h(2))
for k=1:3
series1_l(x,t)=simplify(series1_l(x,t)+u_l(k)*(power(t,k-1)));
series2_l(x,t)=simplify(series2_l(x,t)+v_l(k)*(power(t,k-1)));
series3_l(x,t)=simplify(series3_l(x,t)+h_l(k)*(power(t,k-1)));
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% UPPER VALUE %%%%%%%%%%%%%%%%
for k=1:2
A_up=0;
B_up=0;
C_up=0;
D_up=0;
if k==1
E=1;
else
E=0;
end
for r=1:k
A_up=A_up+u_up(r)*diff(u_up(k-r+1),x,1);
B_up=B_up+u_up(r)*diff(v_up(k-r+1),x,1);
C_up=C_up+u_up(r)*diff(h_up(k-r+1),x,1);
D_up=D_up+h_up(r)*diff(u_up(k-r+1),x,1);
end
u_up(k+1)=simplify(gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-A_up+f*v_up(k)-g*diff(h_up(k),x,1)));
v_up(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-B_up-f*u_up(k)-g*H*E);
h_up(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-C_up-H*v_up(k)-D_up);
end
% t1=simplify(u(2))
% var2 = vpa(t1)_u
% t2=simplify(v(2))
% var = vpa(t2)
% t3=simplify(h(2))
for k=1:3
series1_up(x,t)=simplify(series1_up(x,t)+u_up(k)*(power(t,k-1)));
series2_up(x,t)=simplify(series2_up(x,t)+v_up(k)*(power(t,k-1)));
series3_up(x,t)=simplify(series3_up(x,t)+h_up(k)*(power(t,k-1)));
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%middle value %%%%%%%%%%%%%%
for k=1:2
A_cr=0;
B_cr=0;
C_cr=0;
D_cr=0;
if k==1
E=1;
else
E=0;
end
for r=1:k
A_cr=A_cr+u_cr(r)*diff(u_up(k-r+1),x,1);
B_cr=B_cr+u_cr(r)*diff(v_up(k-r+1),x,1);
C_cr=C_cr+u_cr(r)*diff(h_up(k-r+1),x,1);
D_cr=D_up+h_cr(r)*diff(u_up(k-r+1),x,1);
end
u_cr(k+1)=simplify(gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-A_cr+f*v_up(k)-g*diff(h_cr(k),x,1)));
v_cr(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-B_cr-f*u_cr(k)-g*H*E);
h_cr(k+1)=gamma(((k-1)*alpha)+1)/gamma((alpha*(k+1-1))+1)*(-C_cr-H*v_cr(k)-D_cr);
end
% t1=simplify(u(2))
% var2 = vpa(t1)_u
% t2=simplify(v(2))
% var = vpa(t2)
% t3=simplify(h(2))
for k=1:3
series1_cr(x,t)=simplify(series1_cr(x,t)+u_cr(k)*(power(t,k-1)));
series2_cr(x,t)=simplify(series2_cr(x,t)+v_cr(k)*(power(t,k-1)));
series3_cr(x,t)=simplify(series3_cr(x,t)+h_cr(k)*(power(t,k-1)));
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
C1_l=zeros(1)
C2_l=zeros(1)
C3_l=zeros(1)
C1_up=zeros(1)
C2_up=zeros(1)
C3_up=zeros(1)
C1_cr=zeros(1)
C2_cr=zeros(1)
C3_cr=zeros(1)
row=0;
x=0:0.04:2
t=0:0.002:0.1
for i=1:length(x)
row=row+1;
col=0;
for j=1:length(t)
col=col+1;
C1_l(row,col)=series1_l(x(i),t(j));
C2_l(row,col)=series2_l(x(i),t(j));
C3_l(row,col)=series3_l(x(i),t(j));
%———————————-
C1_up(row,col)=series1_up(x(i),t(j));
C2_up(row,col)=series2_up(x(i),t(j));
C3_up(row,col)=series3_up(x(i),t(j));
%————————————
C1_cr(row,col)=series1_cr(x(i),t(j));
C2_cr(row,col)=series2_cr(x(i),t(j));
C3_cr(row,col)=series3_cr(x(i),t(j));
end
end
%——————————————————–
surf(x,t,C1_l,C1_up,C1_cr)
surf(x,t,C2_l,C2_up,C2_cr)
surf(x,t,C3_l,C3_up,C3_cr)
I want to plot all the graph in the same 3D plot. I dont want different graph using subplot. and I am using MATLAB version 2023a 3 d plot MATLAB Answers — New Questions
using spice models in simlink
Hello,
I have simple spice model defined as described in the attached image.
I want to use this script in the simulink modelling environment.
Is this possible?
Can anyone help with this?
ThanksHello,
I have simple spice model defined as described in the attached image.
I want to use this script in the simulink modelling environment.
Is this possible?
Can anyone help with this?
Thanks Hello,
I have simple spice model defined as described in the attached image.
I want to use this script in the simulink modelling environment.
Is this possible?
Can anyone help with this?
Thanks simulink MATLAB Answers — New Questions
Heatmap inside custom 2d geometry shape
Hi,
I have a specific 2d geometry of an object with multiple fields. I also know the data values for each field. How can I plot a heatmap with my values inside the fileds using the shape of my 2d object. I could get the CAD of the object for example, create a smiple drawing myself, or imitate my 2d shape with blocks or somehow else ….
Thanks.Hi,
I have a specific 2d geometry of an object with multiple fields. I also know the data values for each field. How can I plot a heatmap with my values inside the fileds using the shape of my 2d object. I could get the CAD of the object for example, create a smiple drawing myself, or imitate my 2d shape with blocks or somehow else ….
Thanks. Hi,
I have a specific 2d geometry of an object with multiple fields. I also know the data values for each field. How can I plot a heatmap with my values inside the fileds using the shape of my 2d object. I could get the CAD of the object for example, create a smiple drawing myself, or imitate my 2d shape with blocks or somehow else ….
Thanks. heatmap, plot, 2d, shape, individual, geometry, fields, labels MATLAB Answers — New Questions
sprintf vs. compose performance for large arrays
My understanding is that compose is based on sprintf, yet I’m noticing a huge discrepancy in performance. Consider the following example.
M = 32;
N = 32;
A = randn(M,N);
[m,n] = ndgrid(1:M,1:N);
Nrep = 100;
%% compose and no for loops
t1 = zeros(Nrep,1);
for kr=1:Nrep
tic;
s1 = compose("%d,%d,%.3g",m(:),n(:),A(:));
s1 = reshape(s1,M,N);
t1(kr) = toc;
end
%% sprintf and two nested for loops
t2 = zeros(Nrep,1);
for kr=1:Nrep
s2 = repelem("",M,N);
tic;
for j=1:N
for i=1:M
s2(i,j) = sprintf("%d,%d,%.3g",i,j,A(i,j));
end
end
t2(kr) = toc;
end
%% compose and two nested for loops
t3 = zeros(Nrep,1);
for kr=1:Nrep
tic;
s3 = repelem("",M,N);
for j=1:N
for i=1:M
s3(i,j) = compose("%d,%d,%.3g",i,j,A(i,j));
end
end
t3(kr) = toc;
end
fprintf(" min | mean | maxn");
fprintf("compose, no loops: %.6f | %.6f | %.6fn",min(t1),mean(t1),max(t1));
fprintf("sprintf, 2 loops: %.6f | %.6f | %.6fn",min(t2),mean(t2),max(t2));
fprintf("compose, 2 loops: %.6f | %.6f | %.6fn",min(t3),mean(t3),max(t3));
This code produces the following output on my machine.
min | mean | max
compose, no loops: 1.008151 | 1.035783 | 1.076671
sprintf, 2 loops: 0.004265 | 0.004384 | 0.008990
compose, 2 loops: 1.018900 | 1.050923 | 1.464713
It seems that sprintf is 60x – 70x faster in this example. Any idea why that is?
Here is the output of ver on my machine:
—————————————————————————————–
MATLAB Version: 25.1.0.2943329 (R2025a)
MATLAB License Number:
Operating System: macOS Version: 15.5 Build: 24F74
Java Version: Java 11.0.27+6-LTS with Amazon.com Inc. OpenJDK 64-Bit Server VM mixed mode
—————————————————————————————–My understanding is that compose is based on sprintf, yet I’m noticing a huge discrepancy in performance. Consider the following example.
M = 32;
N = 32;
A = randn(M,N);
[m,n] = ndgrid(1:M,1:N);
Nrep = 100;
%% compose and no for loops
t1 = zeros(Nrep,1);
for kr=1:Nrep
tic;
s1 = compose("%d,%d,%.3g",m(:),n(:),A(:));
s1 = reshape(s1,M,N);
t1(kr) = toc;
end
%% sprintf and two nested for loops
t2 = zeros(Nrep,1);
for kr=1:Nrep
s2 = repelem("",M,N);
tic;
for j=1:N
for i=1:M
s2(i,j) = sprintf("%d,%d,%.3g",i,j,A(i,j));
end
end
t2(kr) = toc;
end
%% compose and two nested for loops
t3 = zeros(Nrep,1);
for kr=1:Nrep
tic;
s3 = repelem("",M,N);
for j=1:N
for i=1:M
s3(i,j) = compose("%d,%d,%.3g",i,j,A(i,j));
end
end
t3(kr) = toc;
end
fprintf(" min | mean | maxn");
fprintf("compose, no loops: %.6f | %.6f | %.6fn",min(t1),mean(t1),max(t1));
fprintf("sprintf, 2 loops: %.6f | %.6f | %.6fn",min(t2),mean(t2),max(t2));
fprintf("compose, 2 loops: %.6f | %.6f | %.6fn",min(t3),mean(t3),max(t3));
This code produces the following output on my machine.
min | mean | max
compose, no loops: 1.008151 | 1.035783 | 1.076671
sprintf, 2 loops: 0.004265 | 0.004384 | 0.008990
compose, 2 loops: 1.018900 | 1.050923 | 1.464713
It seems that sprintf is 60x – 70x faster in this example. Any idea why that is?
Here is the output of ver on my machine:
—————————————————————————————–
MATLAB Version: 25.1.0.2943329 (R2025a)
MATLAB License Number:
Operating System: macOS Version: 15.5 Build: 24F74
Java Version: Java 11.0.27+6-LTS with Amazon.com Inc. OpenJDK 64-Bit Server VM mixed mode
—————————————————————————————– My understanding is that compose is based on sprintf, yet I’m noticing a huge discrepancy in performance. Consider the following example.
M = 32;
N = 32;
A = randn(M,N);
[m,n] = ndgrid(1:M,1:N);
Nrep = 100;
%% compose and no for loops
t1 = zeros(Nrep,1);
for kr=1:Nrep
tic;
s1 = compose("%d,%d,%.3g",m(:),n(:),A(:));
s1 = reshape(s1,M,N);
t1(kr) = toc;
end
%% sprintf and two nested for loops
t2 = zeros(Nrep,1);
for kr=1:Nrep
s2 = repelem("",M,N);
tic;
for j=1:N
for i=1:M
s2(i,j) = sprintf("%d,%d,%.3g",i,j,A(i,j));
end
end
t2(kr) = toc;
end
%% compose and two nested for loops
t3 = zeros(Nrep,1);
for kr=1:Nrep
tic;
s3 = repelem("",M,N);
for j=1:N
for i=1:M
s3(i,j) = compose("%d,%d,%.3g",i,j,A(i,j));
end
end
t3(kr) = toc;
end
fprintf(" min | mean | maxn");
fprintf("compose, no loops: %.6f | %.6f | %.6fn",min(t1),mean(t1),max(t1));
fprintf("sprintf, 2 loops: %.6f | %.6f | %.6fn",min(t2),mean(t2),max(t2));
fprintf("compose, 2 loops: %.6f | %.6f | %.6fn",min(t3),mean(t3),max(t3));
This code produces the following output on my machine.
min | mean | max
compose, no loops: 1.008151 | 1.035783 | 1.076671
sprintf, 2 loops: 0.004265 | 0.004384 | 0.008990
compose, 2 loops: 1.018900 | 1.050923 | 1.464713
It seems that sprintf is 60x – 70x faster in this example. Any idea why that is?
Here is the output of ver on my machine:
—————————————————————————————–
MATLAB Version: 25.1.0.2943329 (R2025a)
MATLAB License Number:
Operating System: macOS Version: 15.5 Build: 24F74
Java Version: Java 11.0.27+6-LTS with Amazon.com Inc. OpenJDK 64-Bit Server VM mixed mode
—————————————————————————————– string, strings, sprintf, compose, text MATLAB Answers — New Questions
I’m sending 170 buffered samples from an ESP32 with MPU6050 to ThingSpeak, but the total entries increase inconsistently with a few samples missing in each interval.
I’m using an ESP32 to collect data from an MPU6050 sensor. The ESP32 buffers 170 samples at a time and uploads them to ThingSpeak using the bulk update API.
However, the data entries received on ThingSpeak are inconsistent. Here’s the pattern of total entries I see on the channel after each upload:
After 1st upload: 170 entries (correct)
After 2nd upload: 338 entries (should be 340)
After 3rd upload: 499 entries (should be 510)
After 4th upload: 657 entries (should be 680)
After 5th upload: 818 entries (should be 850)
As you can see, the expected total should increase by 170 each time, but it’s losing a few samples in each batch, and the number of missing entries increases over time. The losses are not consistent — it seems like 1 or 2 samples get dropped randomly in each interval.
Some things I’ve considered:
The ESP32 is collecting and sending the full buffer (verified via serial debug).
Could it be a payload size limit, HTTP timeout, or formatting issue?
Maybe ThingSpeak is silently dropping malformed lines from the bulk update?
Has anyone else run into this with buffered uploads to ThingSpeak? Any tips for debugging the exact cause of dropped samples?
Thanks for any help!I’m using an ESP32 to collect data from an MPU6050 sensor. The ESP32 buffers 170 samples at a time and uploads them to ThingSpeak using the bulk update API.
However, the data entries received on ThingSpeak are inconsistent. Here’s the pattern of total entries I see on the channel after each upload:
After 1st upload: 170 entries (correct)
After 2nd upload: 338 entries (should be 340)
After 3rd upload: 499 entries (should be 510)
After 4th upload: 657 entries (should be 680)
After 5th upload: 818 entries (should be 850)
As you can see, the expected total should increase by 170 each time, but it’s losing a few samples in each batch, and the number of missing entries increases over time. The losses are not consistent — it seems like 1 or 2 samples get dropped randomly in each interval.
Some things I’ve considered:
The ESP32 is collecting and sending the full buffer (verified via serial debug).
Could it be a payload size limit, HTTP timeout, or formatting issue?
Maybe ThingSpeak is silently dropping malformed lines from the bulk update?
Has anyone else run into this with buffered uploads to ThingSpeak? Any tips for debugging the exact cause of dropped samples?
Thanks for any help! I’m using an ESP32 to collect data from an MPU6050 sensor. The ESP32 buffers 170 samples at a time and uploads them to ThingSpeak using the bulk update API.
However, the data entries received on ThingSpeak are inconsistent. Here’s the pattern of total entries I see on the channel after each upload:
After 1st upload: 170 entries (correct)
After 2nd upload: 338 entries (should be 340)
After 3rd upload: 499 entries (should be 510)
After 4th upload: 657 entries (should be 680)
After 5th upload: 818 entries (should be 850)
As you can see, the expected total should increase by 170 each time, but it’s losing a few samples in each batch, and the number of missing entries increases over time. The losses are not consistent — it seems like 1 or 2 samples get dropped randomly in each interval.
Some things I’ve considered:
The ESP32 is collecting and sending the full buffer (verified via serial debug).
Could it be a payload size limit, HTTP timeout, or formatting issue?
Maybe ThingSpeak is silently dropping malformed lines from the bulk update?
Has anyone else run into this with buffered uploads to ThingSpeak? Any tips for debugging the exact cause of dropped samples?
Thanks for any help! thingspeak, buffer, burst MATLAB Answers — New Questions
defining function which generates 3d array within a class. Different behavior in class versus command window
This code:
lam = @(t) 3*(1 + 0.8*cos(2*pi*t));
A1 = @(t) reshape([lam(t(:).’); lam(t(:).’); zeros(1, numel(t)); lam(t(:).’)], [2, 2, numel(t)]);
tt=0:1/400:1-1/400;
A1stack=A1(tt);
generates a 2x2x400 array. This is what I want to happen. When I embed the code within a class function however, it returns a 2×800 array. How do I fix this?This code:
lam = @(t) 3*(1 + 0.8*cos(2*pi*t));
A1 = @(t) reshape([lam(t(:).’); lam(t(:).’); zeros(1, numel(t)); lam(t(:).’)], [2, 2, numel(t)]);
tt=0:1/400:1-1/400;
A1stack=A1(tt);
generates a 2x2x400 array. This is what I want to happen. When I embed the code within a class function however, it returns a 2×800 array. How do I fix this? This code:
lam = @(t) 3*(1 + 0.8*cos(2*pi*t));
A1 = @(t) reshape([lam(t(:).’); lam(t(:).’); zeros(1, numel(t)); lam(t(:).’)], [2, 2, numel(t)]);
tt=0:1/400:1-1/400;
A1stack=A1(tt);
generates a 2x2x400 array. This is what I want to happen. When I embed the code within a class function however, it returns a 2×800 array. How do I fix this? array function, 3d array, oop MATLAB Answers — New Questions
Word document “Saveas2()” method no longer working
As of the past few days, I seem to be no longer able to save word documents created through matlab. Several months ago I ran the following script which opens an existing word document and saves it as another document.
reportPath = ‘Table_out.docx’;
templateName = append(pwd, ‘Table_Inject_Test.docx’);
actx_word = actxserver(‘Word.Application’);
actx_word.Visible = true;
trace(actx_word.Visible);
wordTemplate = actx_word.Documents.Open(templateName); % Open template
wordTemplate.SaveAs2(reportPath); % Save copy as report
Once I started getting this error in another project, I reverted back to this script to check it wasn’t something I had done within the word template to cause the error.
However when I try to run the same script now, I get the following error:
% Unrecognized method, property, or field ‘SaveAs2’ for class
% ‘Interface.0002096B_0000_0000_C000_000000000046’.
% Error in tableinjecttest2 (line 7)
% wordTemplate.SaveAs2(reportPath); % Save copy as report
Has something changed within the WordInterface object? I see there is a ‘saveobj’ method but that doesn’t seem to save the document either. That just gives me a different error:
% Unable to resolve the name ‘wordParent.saveobj’.
%
% Error in tableInjectTest (line 22)
% wordTemplate.saveobj(reportPath)As of the past few days, I seem to be no longer able to save word documents created through matlab. Several months ago I ran the following script which opens an existing word document and saves it as another document.
reportPath = ‘Table_out.docx’;
templateName = append(pwd, ‘Table_Inject_Test.docx’);
actx_word = actxserver(‘Word.Application’);
actx_word.Visible = true;
trace(actx_word.Visible);
wordTemplate = actx_word.Documents.Open(templateName); % Open template
wordTemplate.SaveAs2(reportPath); % Save copy as report
Once I started getting this error in another project, I reverted back to this script to check it wasn’t something I had done within the word template to cause the error.
However when I try to run the same script now, I get the following error:
% Unrecognized method, property, or field ‘SaveAs2’ for class
% ‘Interface.0002096B_0000_0000_C000_000000000046’.
% Error in tableinjecttest2 (line 7)
% wordTemplate.SaveAs2(reportPath); % Save copy as report
Has something changed within the WordInterface object? I see there is a ‘saveobj’ method but that doesn’t seem to save the document either. That just gives me a different error:
% Unable to resolve the name ‘wordParent.saveobj’.
%
% Error in tableInjectTest (line 22)
% wordTemplate.saveobj(reportPath) As of the past few days, I seem to be no longer able to save word documents created through matlab. Several months ago I ran the following script which opens an existing word document and saves it as another document.
reportPath = ‘Table_out.docx’;
templateName = append(pwd, ‘Table_Inject_Test.docx’);
actx_word = actxserver(‘Word.Application’);
actx_word.Visible = true;
trace(actx_word.Visible);
wordTemplate = actx_word.Documents.Open(templateName); % Open template
wordTemplate.SaveAs2(reportPath); % Save copy as report
Once I started getting this error in another project, I reverted back to this script to check it wasn’t something I had done within the word template to cause the error.
However when I try to run the same script now, I get the following error:
% Unrecognized method, property, or field ‘SaveAs2’ for class
% ‘Interface.0002096B_0000_0000_C000_000000000046’.
% Error in tableinjecttest2 (line 7)
% wordTemplate.SaveAs2(reportPath); % Save copy as report
Has something changed within the WordInterface object? I see there is a ‘saveobj’ method but that doesn’t seem to save the document either. That just gives me a different error:
% Unable to resolve the name ‘wordParent.saveobj’.
%
% Error in tableInjectTest (line 22)
% wordTemplate.saveobj(reportPath) word application, actxserver MATLAB Answers — New Questions
Finding roots of a complex function
Hi,
I’m trying to find the roots of a complex function z=x+iyz in MATLAB. However, it seems that the root-finding routine is missing some roots, which leads to inaccurate results. Could you advise on how I can improve the code to ensure more reliable root detection? I have the two matlab files.
Thanks,
clearvars
close all
% set parameter values
pars.gamma1=0.1093;
pars.alpha3=-0.1104e-2;
pars.K1=6e-12;
pars.d=0.2e-3;
pars.eta1=0.240e-1;
pars.chia=1.219e-6;
pars.alpha=1-pars.alpha3^2/(pars.gamma1*pars.eta1);
pars.Ha=pi*sqrt(pars.K1/pars.chia)/pars.d;
% set lists of u (field) and xi (activity) values
uvals=0:0.05:3;
xivals=-0.3:0.01:0.3;
nu=length(uvals);
nxi=length(xivals);
% initiate arrays for output
taumin=ones(nu,nxi);
wavenummin=ones(nu,nxi);
% start timer
tic
disp(‘Starting u and xi loops’);
% start loop around u values
for i=1:nu
pars.H=uvals(i)*pars.Ha;
% start loop around xi value
for j=1:nxi
xi=xivals(j);
% set initial tau values for root finding, tau is a complex
% variable
tauRvals=-50:0.1:50;
tauIvals=0.1*ones(size(tauRvals));
ntau=length(tauRvals);
% plot equation (projected onto real line) to solve for these values of u and xi
fig1 = figure(1);
y=zeros(size(tauRvals));
for ii=1:ntau
tau=tauRvals(ii);
y(ii) = (pars.H ^ 2 * sin(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.chia * pars.d * tau ^ (0.3e1 / 0.2e1) * sqrt(pars.eta1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) + sin(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.d * pars.gamma1 * sqrt(pars.eta1) * sqrt(tau) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) – 0.2e1 * sqrt(pars.K1) * cos(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.alpha3 * tau ^ 2 * xi + 0.2e1 * sqrt(pars.K1) * cos(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.alpha3 ^ 2 * tau + 0.2e1 * sqrt(pars.K1) * pars.alpha3 * tau ^ 2 * xi – 0.2e1 * sqrt(pars.K1) * pars.alpha3 ^ 2 * tau) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.3e1 / 0.2e1) * (pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) ^ (-0.1e1 / 0.2e1) / pars.alpha3 / sin(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d);
end
plot(tauRvals,y);
hold on
plot(tauRvals,zeros(size(tauRvals)),’-k’);
xline(0);
yline(0);
xlabel(‘tau’);
ylabel(‘tau equation’);
axis([min(tauRvals) max(tauRvals) -1e-2 1e-2]);
drawnow
hold off
% loop around initial tau values for root finding
tausol=zeros(1,ntau);
flag=zeros(1,ntau);
wavenum=zeros(1,ntau);
for k=1:ntau
% set function, options and inital tau value
fun = @(x)rootsolver_complex(x,xi,pars);
options = optimset(‘TolFun’,1e-15,’MaxFunEvals’,1e5,’Maxiter’,1e5,’Display’,’none’);
tauinit=[tauRvals(k),tauIvals(k)];
% find root of tau equation
[x,fval,exitflag,output] = fsolve(fun,tauinit,options);
% save complex tau solution
tausol(k)=complex(x(1),x(2));
% set solve flag (if exitflag>0 the root finder has solved)
flag(k)=(exitflag>0);
% calulate wavenumber (real part of) using equation from Maple
% file
wavenum(k)=real(sqrt((pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) / pars.K1 / pars.eta1 / tau) * pars.d / pi / 0.2e1);
end
tauflag=[tausol’,flag’];
tausol_found=tauflag(flag==1);
tausolR=real(tausol_found);
tauRvals=tauRvals(flag==1);
wavenum=wavenum(flag==1);
% plot real part of tau solution versus initial tau
fig2 = figure(2);
plot(tauRvals,tausolR,’g-‘)
hold on
plot(tauRvals,tauRvals,’k-‘)
xlabel(‘initial $tau$’, ‘Interpreter’,’latex’);
ylabel(‘$tau$ solution’, ‘Interpreter’,’latex’);
hold off
% calculate min value of real part of tau and the wavenumber at
% that min value of tau
taumin(i,j)=min(tausolR);
wavenummins=wavenum(tausolR==min(tausolR));
wavenummin(i,j)=wavenummins(1);
% filled contour plot of minimum tau value (negative tau means instability)
fig3 = figure(3);
[Xi,U] = meshgrid(xivals,uvals);
N=[0:0.1:1];
map = [0.95*(1-N’) 0.95*(1-N’) N’];
contourf(U,Xi,taumin,[-100:10:100])
colormap(map)
colorbar
xlabel(‘Orienting field, $u$’, ‘Interpreter’,’latex’);
ylabel(‘Activity, $xi$ [Pa]’, ‘Interpreter’,’latex’);
%title(‘minimum tau’);
drawnow
% filled contour plot of minimum tau value (negative tau means instability)
fig4 =figure(4);
contourf(U,Xi,wavenummin,10)
colormap(map)
colorbar
xlabel(‘Orienting field, $u$’, ‘Interpreter’,’latex’);
ylabel(‘Activity, $xi$ [Pa]’, ‘Interpreter’,’latex’);
%title(‘wavenumber at minimum tau’);
drawnow
% stability domain in (u,xi) plane
fig5 = figure(5);
S = 25; % size of symbols in pixels
% normalize colouring vector to go from zero to 1
normtau = (taumin>0);
normtau=reshape(normtau,nu*nxi,1);
C = [0.95*(1-normtau) 0.95*(1-normtau) normtau];
scatter(reshape(U,nu*nxi,1),reshape(Xi,nu*nxi,1),S,C,’filled’,’Marker’,’o’)
xlabel(‘Orienting field, $u$’, ‘Interpreter’,’latex’);
ylabel(‘Activity, $xi$ [Pa]’, ‘Interpreter’,’latex’);
title(‘Blue = stable, Yellow = unstable’, ‘Interpreter’,’latex’);
drawnow
end
% display time taken and percentage complete
toc
disp([‘Progress: ‘ num2str(round(100*(i*(j-1)+j)/(nu*nxi))) ‘ % completed’]);
end
function F = rootsolver_complex(x,xi,pars)
% function to provide right-hand-side of the equation for tau
gamma1=pars.gamma1;
alpha3=pars.alpha3;
K1=pars.K1;
d=pars.d;
eta1=pars.eta1;
chia=pars.chia;
alpha=pars.alpha;
H=pars.H;
tau=complex(x(1),x(2));
% equation taken directly from Maple file eq.mw
y = (H ^ 2 * sin(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * chia * d * tau ^ (0.3e1 / 0.2e1) * sqrt(eta1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) + sin(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * d * gamma1 * sqrt(eta1) * sqrt(tau) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) – 0.2e1 * sqrt(K1) * cos(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * alpha3 * tau ^ 2 * xi + 0.2e1 * sqrt(K1) * cos(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * alpha3 ^ 2 * tau + 0.2e1 * sqrt(K1) * alpha3 * tau ^ 2 * xi – 0.2e1 * sqrt(K1) * alpha3 ^ 2 * tau) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.3e1 / 0.2e1) * (H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) ^ (-0.1e1 / 0.2e1) / alpha3 / sin(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d);
F=[real(y),imag(y)];
endHi,
I’m trying to find the roots of a complex function z=x+iyz in MATLAB. However, it seems that the root-finding routine is missing some roots, which leads to inaccurate results. Could you advise on how I can improve the code to ensure more reliable root detection? I have the two matlab files.
Thanks,
clearvars
close all
% set parameter values
pars.gamma1=0.1093;
pars.alpha3=-0.1104e-2;
pars.K1=6e-12;
pars.d=0.2e-3;
pars.eta1=0.240e-1;
pars.chia=1.219e-6;
pars.alpha=1-pars.alpha3^2/(pars.gamma1*pars.eta1);
pars.Ha=pi*sqrt(pars.K1/pars.chia)/pars.d;
% set lists of u (field) and xi (activity) values
uvals=0:0.05:3;
xivals=-0.3:0.01:0.3;
nu=length(uvals);
nxi=length(xivals);
% initiate arrays for output
taumin=ones(nu,nxi);
wavenummin=ones(nu,nxi);
% start timer
tic
disp(‘Starting u and xi loops’);
% start loop around u values
for i=1:nu
pars.H=uvals(i)*pars.Ha;
% start loop around xi value
for j=1:nxi
xi=xivals(j);
% set initial tau values for root finding, tau is a complex
% variable
tauRvals=-50:0.1:50;
tauIvals=0.1*ones(size(tauRvals));
ntau=length(tauRvals);
% plot equation (projected onto real line) to solve for these values of u and xi
fig1 = figure(1);
y=zeros(size(tauRvals));
for ii=1:ntau
tau=tauRvals(ii);
y(ii) = (pars.H ^ 2 * sin(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.chia * pars.d * tau ^ (0.3e1 / 0.2e1) * sqrt(pars.eta1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) + sin(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.d * pars.gamma1 * sqrt(pars.eta1) * sqrt(tau) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) – 0.2e1 * sqrt(pars.K1) * cos(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.alpha3 * tau ^ 2 * xi + 0.2e1 * sqrt(pars.K1) * cos(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.alpha3 ^ 2 * tau + 0.2e1 * sqrt(pars.K1) * pars.alpha3 * tau ^ 2 * xi – 0.2e1 * sqrt(pars.K1) * pars.alpha3 ^ 2 * tau) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.3e1 / 0.2e1) * (pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) ^ (-0.1e1 / 0.2e1) / pars.alpha3 / sin(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d);
end
plot(tauRvals,y);
hold on
plot(tauRvals,zeros(size(tauRvals)),’-k’);
xline(0);
yline(0);
xlabel(‘tau’);
ylabel(‘tau equation’);
axis([min(tauRvals) max(tauRvals) -1e-2 1e-2]);
drawnow
hold off
% loop around initial tau values for root finding
tausol=zeros(1,ntau);
flag=zeros(1,ntau);
wavenum=zeros(1,ntau);
for k=1:ntau
% set function, options and inital tau value
fun = @(x)rootsolver_complex(x,xi,pars);
options = optimset(‘TolFun’,1e-15,’MaxFunEvals’,1e5,’Maxiter’,1e5,’Display’,’none’);
tauinit=[tauRvals(k),tauIvals(k)];
% find root of tau equation
[x,fval,exitflag,output] = fsolve(fun,tauinit,options);
% save complex tau solution
tausol(k)=complex(x(1),x(2));
% set solve flag (if exitflag>0 the root finder has solved)
flag(k)=(exitflag>0);
% calulate wavenumber (real part of) using equation from Maple
% file
wavenum(k)=real(sqrt((pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) / pars.K1 / pars.eta1 / tau) * pars.d / pi / 0.2e1);
end
tauflag=[tausol’,flag’];
tausol_found=tauflag(flag==1);
tausolR=real(tausol_found);
tauRvals=tauRvals(flag==1);
wavenum=wavenum(flag==1);
% plot real part of tau solution versus initial tau
fig2 = figure(2);
plot(tauRvals,tausolR,’g-‘)
hold on
plot(tauRvals,tauRvals,’k-‘)
xlabel(‘initial $tau$’, ‘Interpreter’,’latex’);
ylabel(‘$tau$ solution’, ‘Interpreter’,’latex’);
hold off
% calculate min value of real part of tau and the wavenumber at
% that min value of tau
taumin(i,j)=min(tausolR);
wavenummins=wavenum(tausolR==min(tausolR));
wavenummin(i,j)=wavenummins(1);
% filled contour plot of minimum tau value (negative tau means instability)
fig3 = figure(3);
[Xi,U] = meshgrid(xivals,uvals);
N=[0:0.1:1];
map = [0.95*(1-N’) 0.95*(1-N’) N’];
contourf(U,Xi,taumin,[-100:10:100])
colormap(map)
colorbar
xlabel(‘Orienting field, $u$’, ‘Interpreter’,’latex’);
ylabel(‘Activity, $xi$ [Pa]’, ‘Interpreter’,’latex’);
%title(‘minimum tau’);
drawnow
% filled contour plot of minimum tau value (negative tau means instability)
fig4 =figure(4);
contourf(U,Xi,wavenummin,10)
colormap(map)
colorbar
xlabel(‘Orienting field, $u$’, ‘Interpreter’,’latex’);
ylabel(‘Activity, $xi$ [Pa]’, ‘Interpreter’,’latex’);
%title(‘wavenumber at minimum tau’);
drawnow
% stability domain in (u,xi) plane
fig5 = figure(5);
S = 25; % size of symbols in pixels
% normalize colouring vector to go from zero to 1
normtau = (taumin>0);
normtau=reshape(normtau,nu*nxi,1);
C = [0.95*(1-normtau) 0.95*(1-normtau) normtau];
scatter(reshape(U,nu*nxi,1),reshape(Xi,nu*nxi,1),S,C,’filled’,’Marker’,’o’)
xlabel(‘Orienting field, $u$’, ‘Interpreter’,’latex’);
ylabel(‘Activity, $xi$ [Pa]’, ‘Interpreter’,’latex’);
title(‘Blue = stable, Yellow = unstable’, ‘Interpreter’,’latex’);
drawnow
end
% display time taken and percentage complete
toc
disp([‘Progress: ‘ num2str(round(100*(i*(j-1)+j)/(nu*nxi))) ‘ % completed’]);
end
function F = rootsolver_complex(x,xi,pars)
% function to provide right-hand-side of the equation for tau
gamma1=pars.gamma1;
alpha3=pars.alpha3;
K1=pars.K1;
d=pars.d;
eta1=pars.eta1;
chia=pars.chia;
alpha=pars.alpha;
H=pars.H;
tau=complex(x(1),x(2));
% equation taken directly from Maple file eq.mw
y = (H ^ 2 * sin(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * chia * d * tau ^ (0.3e1 / 0.2e1) * sqrt(eta1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) + sin(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * d * gamma1 * sqrt(eta1) * sqrt(tau) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) – 0.2e1 * sqrt(K1) * cos(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * alpha3 * tau ^ 2 * xi + 0.2e1 * sqrt(K1) * cos(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * alpha3 ^ 2 * tau + 0.2e1 * sqrt(K1) * alpha3 * tau ^ 2 * xi – 0.2e1 * sqrt(K1) * alpha3 ^ 2 * tau) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.3e1 / 0.2e1) * (H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) ^ (-0.1e1 / 0.2e1) / alpha3 / sin(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d);
F=[real(y),imag(y)];
end Hi,
I’m trying to find the roots of a complex function z=x+iyz in MATLAB. However, it seems that the root-finding routine is missing some roots, which leads to inaccurate results. Could you advise on how I can improve the code to ensure more reliable root detection? I have the two matlab files.
Thanks,
clearvars
close all
% set parameter values
pars.gamma1=0.1093;
pars.alpha3=-0.1104e-2;
pars.K1=6e-12;
pars.d=0.2e-3;
pars.eta1=0.240e-1;
pars.chia=1.219e-6;
pars.alpha=1-pars.alpha3^2/(pars.gamma1*pars.eta1);
pars.Ha=pi*sqrt(pars.K1/pars.chia)/pars.d;
% set lists of u (field) and xi (activity) values
uvals=0:0.05:3;
xivals=-0.3:0.01:0.3;
nu=length(uvals);
nxi=length(xivals);
% initiate arrays for output
taumin=ones(nu,nxi);
wavenummin=ones(nu,nxi);
% start timer
tic
disp(‘Starting u and xi loops’);
% start loop around u values
for i=1:nu
pars.H=uvals(i)*pars.Ha;
% start loop around xi value
for j=1:nxi
xi=xivals(j);
% set initial tau values for root finding, tau is a complex
% variable
tauRvals=-50:0.1:50;
tauIvals=0.1*ones(size(tauRvals));
ntau=length(tauRvals);
% plot equation (projected onto real line) to solve for these values of u and xi
fig1 = figure(1);
y=zeros(size(tauRvals));
for ii=1:ntau
tau=tauRvals(ii);
y(ii) = (pars.H ^ 2 * sin(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.chia * pars.d * tau ^ (0.3e1 / 0.2e1) * sqrt(pars.eta1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) + sin(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.d * pars.gamma1 * sqrt(pars.eta1) * sqrt(tau) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) – 0.2e1 * sqrt(pars.K1) * cos(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.alpha3 * tau ^ 2 * xi + 0.2e1 * sqrt(pars.K1) * cos(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d) * pars.alpha3 ^ 2 * tau + 0.2e1 * sqrt(pars.K1) * pars.alpha3 * tau ^ 2 * xi – 0.2e1 * sqrt(pars.K1) * pars.alpha3 ^ 2 * tau) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.3e1 / 0.2e1) * (pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) ^ (-0.1e1 / 0.2e1) / pars.alpha3 / sin(pars.K1 ^ (-0.1e1 / 0.2e1) * pars.eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) * pars.d);
end
plot(tauRvals,y);
hold on
plot(tauRvals,zeros(size(tauRvals)),’-k’);
xline(0);
yline(0);
xlabel(‘tau’);
ylabel(‘tau equation’);
axis([min(tauRvals) max(tauRvals) -1e-2 1e-2]);
drawnow
hold off
% loop around initial tau values for root finding
tausol=zeros(1,ntau);
flag=zeros(1,ntau);
wavenum=zeros(1,ntau);
for k=1:ntau
% set function, options and inital tau value
fun = @(x)rootsolver_complex(x,xi,pars);
options = optimset(‘TolFun’,1e-15,’MaxFunEvals’,1e5,’Maxiter’,1e5,’Display’,’none’);
tauinit=[tauRvals(k),tauIvals(k)];
% find root of tau equation
[x,fval,exitflag,output] = fsolve(fun,tauinit,options);
% save complex tau solution
tausol(k)=complex(x(1),x(2));
% set solve flag (if exitflag>0 the root finder has solved)
flag(k)=(exitflag>0);
% calulate wavenumber (real part of) using equation from Maple
% file
wavenum(k)=real(sqrt((pars.H ^ 2 * pars.chia * pars.eta1 * tau + pars.alpha3 * tau * xi – pars.alpha3 ^ 2 + pars.eta1 * pars.gamma1) / pars.K1 / pars.eta1 / tau) * pars.d / pi / 0.2e1);
end
tauflag=[tausol’,flag’];
tausol_found=tauflag(flag==1);
tausolR=real(tausol_found);
tauRvals=tauRvals(flag==1);
wavenum=wavenum(flag==1);
% plot real part of tau solution versus initial tau
fig2 = figure(2);
plot(tauRvals,tausolR,’g-‘)
hold on
plot(tauRvals,tauRvals,’k-‘)
xlabel(‘initial $tau$’, ‘Interpreter’,’latex’);
ylabel(‘$tau$ solution’, ‘Interpreter’,’latex’);
hold off
% calculate min value of real part of tau and the wavenumber at
% that min value of tau
taumin(i,j)=min(tausolR);
wavenummins=wavenum(tausolR==min(tausolR));
wavenummin(i,j)=wavenummins(1);
% filled contour plot of minimum tau value (negative tau means instability)
fig3 = figure(3);
[Xi,U] = meshgrid(xivals,uvals);
N=[0:0.1:1];
map = [0.95*(1-N’) 0.95*(1-N’) N’];
contourf(U,Xi,taumin,[-100:10:100])
colormap(map)
colorbar
xlabel(‘Orienting field, $u$’, ‘Interpreter’,’latex’);
ylabel(‘Activity, $xi$ [Pa]’, ‘Interpreter’,’latex’);
%title(‘minimum tau’);
drawnow
% filled contour plot of minimum tau value (negative tau means instability)
fig4 =figure(4);
contourf(U,Xi,wavenummin,10)
colormap(map)
colorbar
xlabel(‘Orienting field, $u$’, ‘Interpreter’,’latex’);
ylabel(‘Activity, $xi$ [Pa]’, ‘Interpreter’,’latex’);
%title(‘wavenumber at minimum tau’);
drawnow
% stability domain in (u,xi) plane
fig5 = figure(5);
S = 25; % size of symbols in pixels
% normalize colouring vector to go from zero to 1
normtau = (taumin>0);
normtau=reshape(normtau,nu*nxi,1);
C = [0.95*(1-normtau) 0.95*(1-normtau) normtau];
scatter(reshape(U,nu*nxi,1),reshape(Xi,nu*nxi,1),S,C,’filled’,’Marker’,’o’)
xlabel(‘Orienting field, $u$’, ‘Interpreter’,’latex’);
ylabel(‘Activity, $xi$ [Pa]’, ‘Interpreter’,’latex’);
title(‘Blue = stable, Yellow = unstable’, ‘Interpreter’,’latex’);
drawnow
end
% display time taken and percentage complete
toc
disp([‘Progress: ‘ num2str(round(100*(i*(j-1)+j)/(nu*nxi))) ‘ % completed’]);
end
function F = rootsolver_complex(x,xi,pars)
% function to provide right-hand-side of the equation for tau
gamma1=pars.gamma1;
alpha3=pars.alpha3;
K1=pars.K1;
d=pars.d;
eta1=pars.eta1;
chia=pars.chia;
alpha=pars.alpha;
H=pars.H;
tau=complex(x(1),x(2));
% equation taken directly from Maple file eq.mw
y = (H ^ 2 * sin(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * chia * d * tau ^ (0.3e1 / 0.2e1) * sqrt(eta1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) + sin(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * d * gamma1 * sqrt(eta1) * sqrt(tau) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) – 0.2e1 * sqrt(K1) * cos(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * alpha3 * tau ^ 2 * xi + 0.2e1 * sqrt(K1) * cos(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d) * alpha3 ^ 2 * tau + 0.2e1 * sqrt(K1) * alpha3 * tau ^ 2 * xi – 0.2e1 * sqrt(K1) * alpha3 ^ 2 * tau) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.3e1 / 0.2e1) * (H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) ^ (-0.1e1 / 0.2e1) / alpha3 / sin(K1 ^ (-0.1e1 / 0.2e1) * eta1 ^ (-0.1e1 / 0.2e1) * tau ^ (-0.1e1 / 0.2e1) * sqrt(H ^ 2 * chia * eta1 * tau + alpha3 * tau * xi – alpha3 ^ 2 + eta1 * gamma1) * d);
F=[real(y),imag(y)];
end roots finding, complex finding MATLAB Answers — New Questions
Nonlinear regression of multiple datasets with different functions but wit some shared parameters between datasets
Apologies since I am new to Matlab.
I have a model describing a drug-receptor system with multiple parameters. There are two different observables (binding , activity) for each drug. (I have separate but related equations for predicting the observables). The model has four parameters per drug and four system parameters that are shared for all drugs. I have datasets with a variable number of replicates for each observable (3 binding curves and one response curve per drug). I have data for 12 different drugs. I would appreciate some help setting up global nonlinear regression for all datasets to obtain best estimates of the drug specific and system specific (shared) parameters and their associated 95% confidence intervals. A general example that can handle this type of situation would be helpful to get me started.Apologies since I am new to Matlab.
I have a model describing a drug-receptor system with multiple parameters. There are two different observables (binding , activity) for each drug. (I have separate but related equations for predicting the observables). The model has four parameters per drug and four system parameters that are shared for all drugs. I have datasets with a variable number of replicates for each observable (3 binding curves and one response curve per drug). I have data for 12 different drugs. I would appreciate some help setting up global nonlinear regression for all datasets to obtain best estimates of the drug specific and system specific (shared) parameters and their associated 95% confidence intervals. A general example that can handle this type of situation would be helpful to get me started. Apologies since I am new to Matlab.
I have a model describing a drug-receptor system with multiple parameters. There are two different observables (binding , activity) for each drug. (I have separate but related equations for predicting the observables). The model has four parameters per drug and four system parameters that are shared for all drugs. I have datasets with a variable number of replicates for each observable (3 binding curves and one response curve per drug). I have data for 12 different drugs. I would appreciate some help setting up global nonlinear regression for all datasets to obtain best estimates of the drug specific and system specific (shared) parameters and their associated 95% confidence intervals. A general example that can handle this type of situation would be helpful to get me started. nonlinear regression, shared and not shared parameters, confidence intervals MATLAB Answers — New Questions
Plotting my own validation and loss graph while training a CNN
Within the Deep learning Toolbox it’s possible to enable the training plots within the options. It uses the following code:
Options = trainingOptions(‘Plots’, ‘training-progress’);
This opens an overview of the accuracy and loss while training, like this:
I think these two plots/graphs are amazing for the analysis of the CNN. Now, I’ve ran into an issue regarding enabling the option mentioned above.
This plot reporter causes conflicts when trying to use it in a standalone application using MATLAB Compiler. This conflict can be resolved by simply removing the ‘Plots’, ‘training-progress’ from the options. When removing this, you lose the two plots/graphs aswell.
I’ve tried to see where the measurements are stored during the plotting so I could extract the data and make my own plots/graphs. This was without sucess.
Now my question is: Is it possible to make my own accuracy and loss graphs by somehow extracting the data the program would have used to create the plots with the ‘Plots’ option enabled?
Any insight would help.
Greetings,
ThomasWithin the Deep learning Toolbox it’s possible to enable the training plots within the options. It uses the following code:
Options = trainingOptions(‘Plots’, ‘training-progress’);
This opens an overview of the accuracy and loss while training, like this:
I think these two plots/graphs are amazing for the analysis of the CNN. Now, I’ve ran into an issue regarding enabling the option mentioned above.
This plot reporter causes conflicts when trying to use it in a standalone application using MATLAB Compiler. This conflict can be resolved by simply removing the ‘Plots’, ‘training-progress’ from the options. When removing this, you lose the two plots/graphs aswell.
I’ve tried to see where the measurements are stored during the plotting so I could extract the data and make my own plots/graphs. This was without sucess.
Now my question is: Is it possible to make my own accuracy and loss graphs by somehow extracting the data the program would have used to create the plots with the ‘Plots’ option enabled?
Any insight would help.
Greetings,
Thomas Within the Deep learning Toolbox it’s possible to enable the training plots within the options. It uses the following code:
Options = trainingOptions(‘Plots’, ‘training-progress’);
This opens an overview of the accuracy and loss while training, like this:
I think these two plots/graphs are amazing for the analysis of the CNN. Now, I’ve ran into an issue regarding enabling the option mentioned above.
This plot reporter causes conflicts when trying to use it in a standalone application using MATLAB Compiler. This conflict can be resolved by simply removing the ‘Plots’, ‘training-progress’ from the options. When removing this, you lose the two plots/graphs aswell.
I’ve tried to see where the measurements are stored during the plotting so I could extract the data and make my own plots/graphs. This was without sucess.
Now my question is: Is it possible to make my own accuracy and loss graphs by somehow extracting the data the program would have used to create the plots with the ‘Plots’ option enabled?
Any insight would help.
Greetings,
Thomas convolutional neural network, deep learning toolbox, accuracy, loss, plots, extract data, training-progress MATLAB Answers — New Questions
Unrecognized function or variable ‘yalmip’.
matlab should known sdpvar in the code becuse if i used help it give me the meaning of it but it said instead
Unrecognized function or variable ‘yalmip’.
Error in sdpvar (line 518)
[mt,variabletype,hashed_monoms,current_hash] = yalmip(‘monomtable’);
% Define the partially specified Euclidean distance matrix
D = [0 1 0 3; 1 0 2 0; 0 2 0 0; 3 0 0 0];
% Define the size of the matrix
n = size(D, 1);
% Define the optimization variables
X = sdpvar(n, n);
% Set up the optimization problem
Constraints = [X == semidefinite(n), diag(X) == zeros(n, 1), X >= 0];
Objective = sum(sum((X – D).^2));
Options = sdpsettings(‘solver’, ‘mosek’);
% Solve the optimization problem
optimize(Constraints, Objective, Options);
% Extract the completed Euclidean distance matrix
D_complete = value(X);matlab should known sdpvar in the code becuse if i used help it give me the meaning of it but it said instead
Unrecognized function or variable ‘yalmip’.
Error in sdpvar (line 518)
[mt,variabletype,hashed_monoms,current_hash] = yalmip(‘monomtable’);
% Define the partially specified Euclidean distance matrix
D = [0 1 0 3; 1 0 2 0; 0 2 0 0; 3 0 0 0];
% Define the size of the matrix
n = size(D, 1);
% Define the optimization variables
X = sdpvar(n, n);
% Set up the optimization problem
Constraints = [X == semidefinite(n), diag(X) == zeros(n, 1), X >= 0];
Objective = sum(sum((X – D).^2));
Options = sdpsettings(‘solver’, ‘mosek’);
% Solve the optimization problem
optimize(Constraints, Objective, Options);
% Extract the completed Euclidean distance matrix
D_complete = value(X); matlab should known sdpvar in the code becuse if i used help it give me the meaning of it but it said instead
Unrecognized function or variable ‘yalmip’.
Error in sdpvar (line 518)
[mt,variabletype,hashed_monoms,current_hash] = yalmip(‘monomtable’);
% Define the partially specified Euclidean distance matrix
D = [0 1 0 3; 1 0 2 0; 0 2 0 0; 3 0 0 0];
% Define the size of the matrix
n = size(D, 1);
% Define the optimization variables
X = sdpvar(n, n);
% Set up the optimization problem
Constraints = [X == semidefinite(n), diag(X) == zeros(n, 1), X >= 0];
Objective = sum(sum((X – D).^2));
Options = sdpsettings(‘solver’, ‘mosek’);
% Solve the optimization problem
optimize(Constraints, Objective, Options);
% Extract the completed Euclidean distance matrix
D_complete = value(X); unrecognized function, sdpvar, yalmip MATLAB Answers — New Questions
Monitor Gamma Calibration (manual and grayscale)
Hello everyone,
Would like your help with monitor gamma calibration:
Does anybody have a script that helps doing gamma calibration manually with a photometer (grayscale, not colors), and creates a gamma table which can be used in experiments?
I created a script with GPT, did a calibration with a photometer and got gamma value of 1.98. when I use the gamma table in my experiment, the the monitor seems much brighter and the stimuli are harder to distinct (it seems like the contrast decreased). Does it make sense?
Thanks in advance :)Hello everyone,
Would like your help with monitor gamma calibration:
Does anybody have a script that helps doing gamma calibration manually with a photometer (grayscale, not colors), and creates a gamma table which can be used in experiments?
I created a script with GPT, did a calibration with a photometer and got gamma value of 1.98. when I use the gamma table in my experiment, the the monitor seems much brighter and the stimuli are harder to distinct (it seems like the contrast decreased). Does it make sense?
Thanks in advance 🙂 Hello everyone,
Would like your help with monitor gamma calibration:
Does anybody have a script that helps doing gamma calibration manually with a photometer (grayscale, not colors), and creates a gamma table which can be used in experiments?
I created a script with GPT, did a calibration with a photometer and got gamma value of 1.98. when I use the gamma table in my experiment, the the monitor seems much brighter and the stimuli are harder to distinct (it seems like the contrast decreased). Does it make sense?
Thanks in advance 🙂 gamma correction, photometer, cognitive experiments, perception, contrast, rgb MATLAB Answers — New Questions
Announcing comprehensive sovereign solutions empowering European organizations
Today, we are taking the next step in strengthening our European Digital Commitments to empower our customers with greater choice, more control over their data privacy and the most robust digital resilience we have ever offered. Building on our 42-year history as a company in Europe, we are expanding our efforts with Microsoft Sovereign Cloud. This offer spans both public cloud and private digital infrastructure, ensuring our customers can choose the right balance of control, compliance and capability for their needs.
With this expanded offering we are announcing Data Guardian for European operations, External Key Management for customer-controlled encryption, Regulated Environment Management for simplified configuration and Microsoft 365 Local for critical productivity services in private cloud environments.
This brings together comprehensive productivity, security and cloud solutions designed to enable European organizations to grow, compete and lead on their own terms and with more control than ever before across Sovereign Public Cloud, Sovereign Private Cloud and National Partner Clouds.
Building on our experience delivering sovereignty solutions that meet the needs of highly regulated customers and government agencies, our Sovereign Public Cloud is an evolution and expansion of the Microsoft Cloud for Sovereignty and will be offered across all existing European datacenter regions, for all European customers, across enterprise services such as Microsoft Azure, Microsoft 365, Microsoft Security and Power Platform. Sovereign Public Cloud ensures customer data stays in Europe, under European Law, with operations and access controlled by European personnel, and encryption is under full control of customers. This is enabled for all customer workloads running in our European datacenter regions requiring no migration.
Microsoft’s new Sovereign Private Cloud will support critical collaboration, communication and virtualization services workloads on Azure Local. This solution now integrates Microsoft 365 Local and our security platform with Azure Local, providing consistent capabilities for hybrid or air-gapped environments to meet resiliency and business continuity requirements.
In France and Germany, our National Partner Clouds offer comprehensive capabilities of Microsoft 365 and Microsoft Azure in an independently owned and operated environment. In France, we have an agreement with Bleu, a joint venture between Orange and Capgemini, for Bleu to operate a “cloud de confiance” for the French public sector, critical infrastructure providers and essential services providers that is designed to meet SecNumCloud requirements. In Germany, we have an agreement with Delos Cloud, an SAP subsidiary, for Delos Cloud to operate a sovereign cloud for the German public sector that is designed to meet the German government’s Cloud Platform Requirements.
Across our Sovereign Public Cloud, Sovereign Private Cloud and support for National Partner Clouds, Microsoft Sovereign Cloud offers the most comprehensive set of sovereignty solutions in the industry for integrated productivity, security and cloud.
Sovereign Public Cloud for all Microsoft Cloud customers in Europe
Many technology providers have approached sovereignty as niche requirements for a unique set of customers that require a specific deployment approach that at times is at odds with the economics and innovation of public cloud systems. This often requires running duplicate systems and teams, migrating to separate environments and limiting access to cutting-edge technologies like AI. However, Microsoft’s Sovereign Public Cloud builds an evolving set of sovereign capabilities that can be configured to meet specific needs without sacrificing functionality or requiring migration to specialized datacenters. With Microsoft’s Sovereign Public Cloud currently in preview and set to be generally available in all European cloud regions later this year, we will introduce new features and solutions that reinforce this vision.
Announcing Data Guardian
Our EU Data Boundary already provides an industry-leading commitment to store and process your data on infrastructure located in Europe. Data Guardian will add an additional level of assurance by ensuring that only Microsoft personnel residing in Europe control remote access to these systems. Data Guardian adds additional human and technical oversight whenever engineers outside of Europe need access. All remote access by Microsoft engineers to the systems that store and process your data in Europe is approved and monitored by European resident personnel in real time and will be logged in a tamper-evident ledger.
Announcing External Key Management to extend Azure Managed HSM
Encryption under the full control of customers provides an additional guarantee of data protection. With external key management, customers can connect Azure to keys stored on their own Hardware Security Module (HSM) on-premises or hosted by a trusted third party. We’re working with major HSM manufacturers such as Futurex, Thales and Utimaco to ensure their support.
Announcing Regulated Environment Management
The Regulated Environment Management service will allow customers to easily manage all these features in one place (for instance, configuring Data Guardian policies or reviewing access log entries). Regulated Environment Management will be at the center of the customer experience for configuring, deploying and monitoring workloads in support of sovereign operations. Together, these tools will be at the center of the customer experience for configuring, deploying and monitoring workloads in the Sovereign Public Cloud.
Sovereign Private Cloud with Azure Local and Microsoft 365 Local
While strengthening sovereign controls in public cloud environments is critical, we also understand that some scenarios require certain workloads be run in a physical environment under full customer control to support business continuity risk mitigation. Azure Local delivers Microsoft cloud services in customer locations, enabling organizations to meet specific data residency and sovereignty requirements. It includes core Azure capabilities — such as compute, storage, networking and virtualization services — while providing a consistent management and developer experience. Azure Local is ideal for delivering services closer to where data is generated or regulated, whether in-country, on-premises or in partner-operated datacenters. Microsoft’s Sovereign Private Cloud solution is in preview today and will be generally available later this year.
Announcing Microsoft 365 Local
Microsoft 365 Local provides customers with additional choice by bringing together Microsoft’s productivity server software into an Azure Local environment that can run entirely in a customer’s own datacenter.
This provides a simplified deployment and management framework for organizations to run Microsoft’s trusted productivity servers in environments they fully control. Built on our validated reference architecture and powered by Azure Local, Microsoft 365 Local enables customers to deploy Microsoft productivity workloads like Exchange Server and SharePoint Server in their own datacenters or sovereign cloud environments — with full control on security, compliance and governance.
Private Sovereign Cloud is designed for governments, critical industries and regulated sectors that need to meet the highest standards of data residency, operational autonomy and disconnected access.
Building a sovereign cloud and AI partner ecosystem for Europe
To support European customers in implementing and operating sovereign solutions, we are also excited to preview a new Microsoft Sovereign Cloud specialization in the Microsoft AI Cloud Partner Program. This specialization will provide our European customers the ability to identify Partners who have differentiated themselves based on their demonstrated capabilities in supporting their Sovereign Cloud ambitions on Microsoft technology. Our preview partners include Accenture, Arvato Systems, Atea, Atos, Crayon, Capgemini, Dell Technologies, IBM, Inspark, Infosys, Lenovo, Leonardo, NTT Data, Orange, Telefonica and Vodafone.
“The launch of Microsoft Sovereign Cloud marks a pivotal moment in empowering European institutions and industries with the control, compliance and innovation they need to thrive in today’s digital economy,” said Aiman Ezzat, CEO of Capgemini Group.
“As a shareholder of Bleu, we have already set up a National Partner Cloud in France in order to deliver Microsoft technologies in a sovereign environment that respects the French State requirements. With decades of experience in Microsoft technologies and deep expertise in regulated sectors, we are uniquely positioned to help our clients harness the full power of Microsoft’s sovereign public and private cloud solutions. Together, we are enabling a trusted digital future for Europe.”
Delivering on our digital commitments to Europe
Together, Microsoft Sovereign Cloud is grounded in our European Digital Commitments and offers the best mix of choice, control and resilience for European customers. Microsoft is proud to offer the broadest set of sovereignty solutions available on the market today and we will constantly look for new ways to ensure our European customers have the options and assurances they need to operate with confidence.
In a time of geopolitical volatility, we are committed to providing digital stability. With each step we take in this journey, we invite open dialogues with our customers, policymakers and regulators as we continue to innovate.
The post Announcing comprehensive sovereign solutions empowering European organizations appeared first on The Official Microsoft Blog.
Today, we are taking the next step in strengthening our European Digital Commitments to empower our customers with greater choice, more control over their data privacy and the most robust digital resilience we have ever offered. Building on our 42-year history as a company in Europe, we are expanding our efforts with Microsoft Sovereign Cloud….
The post Announcing comprehensive sovereign solutions empowering European organizations appeared first on The Official Microsoft Blog.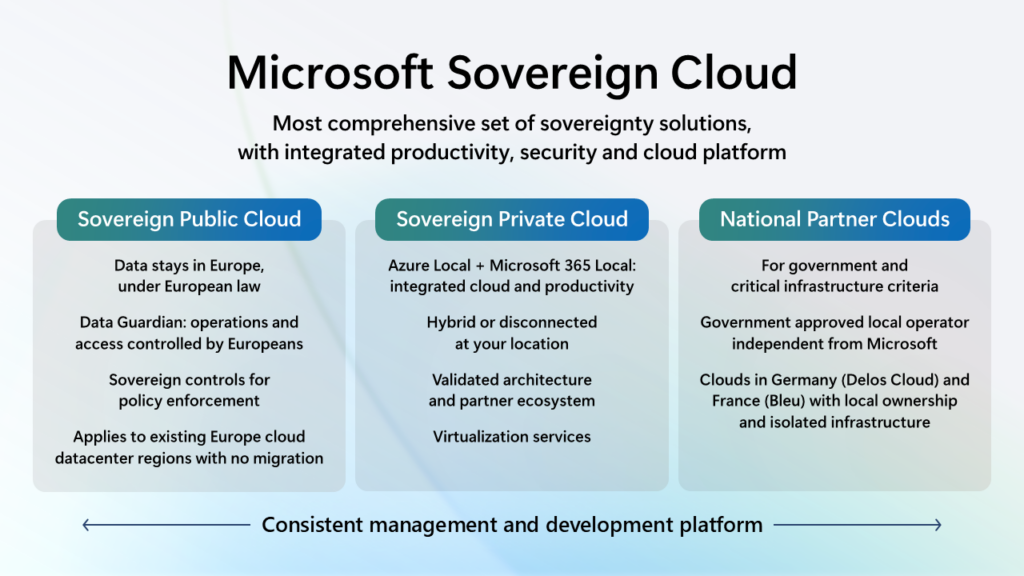 Read More
Read More
Can’t Share Heat with Pipe (G)
Hi,
I am in intership and I would like in Simscape to simulate an air duct heated by a heat source. For this, a started with a block Pipe (G) with a lenght of 5m, an area of 0.5 m^2 and I used the formula for the hydraulic diametre D_h=4A/P. Then I put a block Reservoir (G) on both side of the pipe with an ambiant temperature (25°C) and 1 atm and I put also a flow source with a volumetric flow of 5 m^3/s.
On the port H, I started with a temperature source (40°C) and a convective block with an area of 1.7 m^2 and a heat transfert coefficient of 20 W/(m^2.K).
When I launch my simulation, the temperature of the air remain at 25°C and the temperature of the thermal part drop to 26.7°C. I have a heat flow of 452.6 W from thermal part to gas part so the air have to be heated but it’s not the case.
Can you please help me ? Thank you !Hi,
I am in intership and I would like in Simscape to simulate an air duct heated by a heat source. For this, a started with a block Pipe (G) with a lenght of 5m, an area of 0.5 m^2 and I used the formula for the hydraulic diametre D_h=4A/P. Then I put a block Reservoir (G) on both side of the pipe with an ambiant temperature (25°C) and 1 atm and I put also a flow source with a volumetric flow of 5 m^3/s.
On the port H, I started with a temperature source (40°C) and a convective block with an area of 1.7 m^2 and a heat transfert coefficient of 20 W/(m^2.K).
When I launch my simulation, the temperature of the air remain at 25°C and the temperature of the thermal part drop to 26.7°C. I have a heat flow of 452.6 W from thermal part to gas part so the air have to be heated but it’s not the case.
Can you please help me ? Thank you ! Hi,
I am in intership and I would like in Simscape to simulate an air duct heated by a heat source. For this, a started with a block Pipe (G) with a lenght of 5m, an area of 0.5 m^2 and I used the formula for the hydraulic diametre D_h=4A/P. Then I put a block Reservoir (G) on both side of the pipe with an ambiant temperature (25°C) and 1 atm and I put also a flow source with a volumetric flow of 5 m^3/s.
On the port H, I started with a temperature source (40°C) and a convective block with an area of 1.7 m^2 and a heat transfert coefficient of 20 W/(m^2.K).
When I launch my simulation, the temperature of the air remain at 25°C and the temperature of the thermal part drop to 26.7°C. I have a heat flow of 452.6 W from thermal part to gas part so the air have to be heated but it’s not the case.
Can you please help me ? Thank you ! simscape, urgent MATLAB Answers — New Questions
How to use Simulink for model-based development of TC3XX series development boards
I want to use Simulink for model-based development of the TC3XX series development board, but I am not sure about the development process and toolchain? Is there a free toolchain available for me to use? Do you have a detailed development tutorial? Excuse me, big shotsI want to use Simulink for model-based development of the TC3XX series development board, but I am not sure about the development process and toolchain? Is there a free toolchain available for me to use? Do you have a detailed development tutorial? Excuse me, big shots I want to use Simulink for model-based development of the TC3XX series development board, but I am not sure about the development process and toolchain? Is there a free toolchain available for me to use? Do you have a detailed development tutorial? Excuse me, big shots infineon tc3xx simulink;mbd MATLAB Answers — New Questions
Dynamic filepath for sftp not allowed
Home="/folder1/folder2";
Scanname="Exp04_Sample1";
Sftp_path=strcat(Home,"/",Scanname);
fileInfo=dir(s,Sftp_path);
Why does the above not work?
But
fileInfo=dir(s,"/folder1/folder2/Exp04_Sample1");
will work?Home="/folder1/folder2";
Scanname="Exp04_Sample1";
Sftp_path=strcat(Home,"/",Scanname);
fileInfo=dir(s,Sftp_path);
Why does the above not work?
But
fileInfo=dir(s,"/folder1/folder2/Exp04_Sample1");
will work? Home="/folder1/folder2";
Scanname="Exp04_Sample1";
Sftp_path=strcat(Home,"/",Scanname);
fileInfo=dir(s,Sftp_path);
Why does the above not work?
But
fileInfo=dir(s,"/folder1/folder2/Exp04_Sample1");
will work? sftp MATLAB Answers — New Questions
Shading an area between two curves symbolically
I want to shade the area under the line y=2 and above the curve y=1+cos(x) from 0 to pi. I’m having trouble doing this. This is my code:
dy1 = 1+cos(x);
dy2 = 2;
fplot(dy1, [0 pi]); hold on;
fplot(dy2, [0 pi]);
patch([x fliplr(x)], [dy2 fliplr(dy1)], ‘g’);
hold off;I want to shade the area under the line y=2 and above the curve y=1+cos(x) from 0 to pi. I’m having trouble doing this. This is my code:
dy1 = 1+cos(x);
dy2 = 2;
fplot(dy1, [0 pi]); hold on;
fplot(dy2, [0 pi]);
patch([x fliplr(x)], [dy2 fliplr(dy1)], ‘g’);
hold off; I want to shade the area under the line y=2 and above the curve y=1+cos(x) from 0 to pi. I’m having trouble doing this. This is my code:
dy1 = 1+cos(x);
dy2 = 2;
fplot(dy1, [0 pi]); hold on;
fplot(dy2, [0 pi]);
patch([x fliplr(x)], [dy2 fliplr(dy1)], ‘g’);
hold off; fliplr, symbolic, fplot MATLAB Answers — New Questions
How can one switch between locations of same namespace in one file system?
Kind reader,
I work with a namespace to structure function families. The development folder is a git checkout and I managed to write an installer for the namespace. So the relesed state of the namespace is in the Program Data folder and the path is added in Matlab.
Questions: As boths namespace folders path are now added to Matlab
How can I choose between the two?
How can I activated/deactivate one of them?
———————- File system depiction ——————————–
– C:gitClone/+myNamespace -C:ProgramData/myNamespace
– /+awsomeFunctions – /+awsomeFunctions
– /+helpfulFunctions – /+helpfulFunctions
—————————————————————————–
As I provide a release of the namespace to co-workers I have to be able to check the installation on my development computer. I read about package creation in Matlab (mpmcreate – Create package – MATLAB) but that does not work for namespaces.
Every hint, comment, and thought about this issue is highly appreciated 🙂
Environement: Windows with Matlab 2024bKind reader,
I work with a namespace to structure function families. The development folder is a git checkout and I managed to write an installer for the namespace. So the relesed state of the namespace is in the Program Data folder and the path is added in Matlab.
Questions: As boths namespace folders path are now added to Matlab
How can I choose between the two?
How can I activated/deactivate one of them?
———————- File system depiction ——————————–
– C:gitClone/+myNamespace -C:ProgramData/myNamespace
– /+awsomeFunctions – /+awsomeFunctions
– /+helpfulFunctions – /+helpfulFunctions
—————————————————————————–
As I provide a release of the namespace to co-workers I have to be able to check the installation on my development computer. I read about package creation in Matlab (mpmcreate – Create package – MATLAB) but that does not work for namespaces.
Every hint, comment, and thought about this issue is highly appreciated 🙂
Environement: Windows with Matlab 2024b Kind reader,
I work with a namespace to structure function families. The development folder is a git checkout and I managed to write an installer for the namespace. So the relesed state of the namespace is in the Program Data folder and the path is added in Matlab.
Questions: As boths namespace folders path are now added to Matlab
How can I choose between the two?
How can I activated/deactivate one of them?
———————- File system depiction ——————————–
– C:gitClone/+myNamespace -C:ProgramData/myNamespace
– /+awsomeFunctions – /+awsomeFunctions
– /+helpfulFunctions – /+helpfulFunctions
—————————————————————————–
As I provide a release of the namespace to co-workers I have to be able to check the installation on my development computer. I read about package creation in Matlab (mpmcreate – Create package – MATLAB) but that does not work for namespaces.
Every hint, comment, and thought about this issue is highly appreciated 🙂
Environement: Windows with Matlab 2024b namespace, pathhandling MATLAB Answers — New Questions
How to input an image from the user in simulink?
I have a MATLAB script that I want to run in Simulink (by converting it into a MATLAB function). The code I want to run goes like this:
In this script, the input can be taken from the uigetfile(). How can I take the input from the user in Simulink and pass it in this function so that it can be converted into its binary bits?
Just, for more clarification, the block to be used is,
For the input "u", I want to take image input from the user and on the "y", I would like to have it converted into its binary bits (500*500*3 uint8, in my case).
ThanksI have a MATLAB script that I want to run in Simulink (by converting it into a MATLAB function). The code I want to run goes like this:
In this script, the input can be taken from the uigetfile(). How can I take the input from the user in Simulink and pass it in this function so that it can be converted into its binary bits?
Just, for more clarification, the block to be used is,
For the input "u", I want to take image input from the user and on the "y", I would like to have it converted into its binary bits (500*500*3 uint8, in my case).
Thanks I have a MATLAB script that I want to run in Simulink (by converting it into a MATLAB function). The code I want to run goes like this:
In this script, the input can be taken from the uigetfile(). How can I take the input from the user in Simulink and pass it in this function so that it can be converted into its binary bits?
Just, for more clarification, the block to be used is,
For the input "u", I want to take image input from the user and on the "y", I would like to have it converted into its binary bits (500*500*3 uint8, in my case).
Thanks matlab, simulink, image processing MATLAB Answers — New Questions











